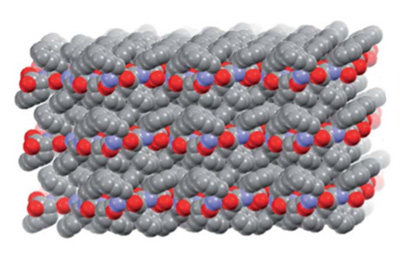
By mixing a small peptide with equal amounts of its mirror image, a team of scientists at UC Santa Cruz has created an unusual protein structure known as a “rippled beta sheet” and obtained images of it using x-ray crystallography. They reported their findings in a paper published December 8 in Chemical Science.
The rippled sheet is a distinctive variation on the pleated beta sheet, which is a well-known structural motif found in thousands of proteins, including important disease-related proteins. Linus Pauling and Robert Corey described the rippled beta sheet in 1953, two years after introducing the concept of the pleated beta sheet.
While the pleated beta sheet (often called simply the beta sheet) quickly became a textbook example of a common protein structure, the rippled sheet has languished in obscurity as a rarely studied and largely theoretical structure. Previous studies have found experimental evidence of rippled sheet formation, but none using x-ray crystallography, which is the gold standard for determining protein structures.
“Now, for the first time, we have the crystal structure of a rippled sheet, which is like a snapshot of it, and the structure closely matches the predictions of Pauling and Corey,” said Jevgenij Raskatov, associate professor of chemistry and biochemistry at UC Santa Cruz and corresponding author of the paper.
“The rippled sheet paradigm may have significance for both materials research and biomedical applications, and having the crystal structure is important for the rational design of rippled sheet materials,” Raskatov noted.
Proteins consist of long chains of amino acids folded into complex three-dimensional shapes that enable them to carry out a huge variety of functions in all living things. A pleated beta sheet is composed of linear strands (called beta strands) bonded together side by side to form a 2-dimensional sheet-like structure. A rippled beta sheet is similar except that alternate strands are mirror images of each other.
The amino acids that make up proteins can have either a “left-handed” (L) or “right-handed” (D) orientation in the arrangement of their atoms—the same in all respects but mirror images, like left and right hands. All natural proteins are made with left-handed amino acids, but synthetic proteins can be made with either L or D amino acids.
In the new study, the researchers used mirror-image forms of triphenylalanine, a short peptide consisting of three phenylalanine amino acids. When mixed in equal amounts, the mirror-image peptides joined in pairs, which then packed together into herringbone layer structures.
“They pack together to form a crystal, so we could use x-ray crystallography to see that rippled sheet structure,” said coauthor Timothy Johnstone, assistant professor of chemistry and biochemistry. “It’s a highly enabling discovery that opens up new avenues for exploration, because it gives us a new building block, or a new way to put building blocks together, for creating novel polypeptide structures with desirable properties.”
Having determined the crystal structure, the researchers then searched the Protein Data Bank, an online archive of structural data, for other proteins involving mirror-image peptides. They found three additional crystal structures containing rippled sheets that had not been recognized when the structures were originally analyzed.
The co-first authors of the paper are Ariel Kuhn, a Ph.D. student in Raskatov’s lab, and Beatriz Ehlke, a Ph.D. student in the lab of coauthor Scott Oliver, professor of chemistry and biochemistry.
“It was a great collaborative effort between the three labs, as well as demonstrating the incredible capabilities of our new single crystal XRD instrument for x-ray crystallography,” Kuhn said.
This work was supported by the National Institutes of Health and the National Science Foundation.