Campus News
Long noncoding RNA identified as a key regulator of inflammation
New findings add to growing evidence that previously dismissed “junk DNA” actually produces RNA molecules with important regulatory functions.
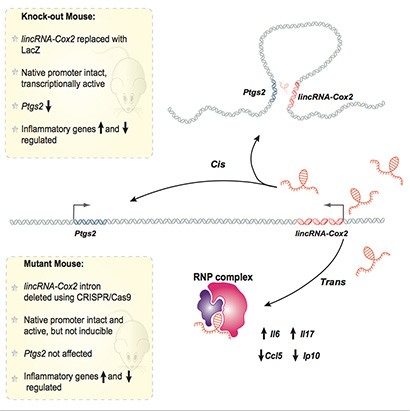

Scientists have identified an RNA molecule with broad powers to regulate the body’s inflammatory response to infection and injury. Called lincRNA-Cox2, it belongs to a recently discovered, highly abundant class of RNAs whose functions are only beginning to be understood.
The sequencing of the human genome revealed that only a small fraction of the DNA in our chromosomes comprises genes that encode instructions for making proteins. Those genes are transcribed into messenger RNA, which directs the synthesis of proteins that carry out various functions in the cell. The rest of the genome, about 98 percent of it, was sometimes referred to as the “dark matter” of the genome or dismissed as “junk DNA.”
In the past decade, however, new RNA sequencing technologies have revealed that much of the genome is transcribed into noncoding RNA molecules of various types. Long intergenic noncoding RNA (lincRNA) is the largest class of these RNAs.
“We now know of about 16,000 long noncoding RNAs, about as many as there are protein-coding genes, but we know the functions of less than one percent of them,” said Susan Carpenter, an assistant professor of molecular, cell and developmental biology at UC Santa Cruz.
Carpenter’s lab is interested in how lincRNAs control the processes involved in inflammation. The new study, published November 6 in Cell Reports, shows that lincRNA-Cox2 functions in several different ways to regulate the activity of genes involved in inflammation and other immune system responses.
Inflammatory gene
Inflammation is a normal part of the body’s response to infection and injury, but unresolved or chronic inflammation is associated with a wide range of diseases. LincRNA-Cox2 was named for its proximity in the genome to a gene called Cox2, which Carpenter calls “the most important inflammatory gene in the body.” Aspirin, ibuprofen, and other nonsteroidal anti-inflammatory drugs (NSAIDs) reduce inflammation by inhibiting the Cox2 enzyme encoded by this gene.
One of the major findings of the new study is that lincRNA-Cox2 regulates the activity of this neighboring gene, boosting production of the enzyme. Carpenter’s team found that levels of the Cox2 enzyme were 70 to 80 percent lower than normal in mice lacking the lincRNA-Cox2 gene.
But Cox2 is not the only gene regulated by lincRNA-Cox2. It also influences the expression of genes scattered all across the genome, and these other genes are important in the innate immune response to infections. LincRNA-Cox2 can inhibit some genes and enhance the expression of others. Previous work by Carpenter’s lab and others had shown these effects in cell cultures, but the new study used genetically altered mice to tease out the effects of lincRNA-Cox2 on the neighboring Cox2 gene and on other immune system genes in live animals.
“We’ve now made animal models that show lincRNA-Cox2 is important in the whole organism, and this is probably one of the first examples to show a lincRNA controls not only a neighboring gene but other genes as well,” Carpenter said.
Viral infections
One of the functions of lincRNA-Cox2 is to inhibit the expression of a number of genes that are important in the innate immune response to viral infections. “You want those genes to turn on when you have an infection, but when they get turned on in the absence of infection it’s associated with autoimmune diseases like lupus,” Carpenter said.
Carpenter’s team also used the mouse models to investigate the expression of lincRNA-Cox2 in different tissues. They found, for example, that it is highly active in lung tissue, which suggests it may play a role in respiratory infections. In future work, the researchers plan to take a closer look at the activity of lincRNA-Cox2 in specific cell types.
“This field is so new, and there was a lot of skepticism at first, but we’re now seeing that these lincRNAs are extremely important,” Carpenter said. “It takes less energy for the cell to make an RNA than it does to make a protein, so it makes sense that a lot of regulatory molecules are RNAs.”
Medical researchers are eager to understand the functions of these noncoding RNA molecules. With the explosive growth of genome sequencing, biomedical scientists have performed many genome-wide association studies (GWAS) to identify genetic variants associated with diseases. But most of the disease-associated genetic variants found in these studies are not located in protein-coding genes.
“It turns out that 97 percent of them are in noncoding regions of the genome,” Carpenter said. “That’s one reason there is so much interest in these noncoding RNA genes.”
In addition to Carpenter, the coauthors of the paper include first authors Roland Elling at the University of Massachusetts Medical School, Worcester, and Elektra Robinson at UC Santa Cruz, as well as researchers at Harvard University, University of Pennsylvania, UC San Francisco, and University of Colorado, Boulder. This work was funded by the National Institutes of Health.