Campus News
Study finds convergent evolution of gene regulation in humans and mice
Molecular biologists have found evidence of convergent evolution in an important mechanism of gene regulation in humans and mice.
Organisms that aren’t closely related may evolve similar traits as they adapt to similar challenges. It’s called convergent evolution, and familiar examples include the wings of birds, bats, and insects, and echolocation in bats and dolphins. Now, molecular biologists have found evidence of convergent evolution in an important mechanism of gene regulation in humans and mice.
The new study, published January 15 in Proceedings of the National Academy of Sciences, was led by researchers at UC Santa Cruz and the University of Rochester Medical Center. They described a complex system that regulates the same genes in the same way in both species, yet evolved independently in the two lineages.
In both cases, the regulatory system involves noncoding RNA (sequences not translated into protein molecules) with origins in DNA segments randomly inserted into the genome by “jumping genes” (retrotransposons).
“This study highlights the importance of noncoding RNA and transposable elements in the regulation of gene expression and in the evolution of gene expression networks in mammalian genomes,” said coauthor Manuel Ares, professor of molecular, cell, and developmental biology at UC Santa Cruz.
Only about 2 percent of the human genome is copied into messenger RNA molecules to code for the proteins that run the main processes in all cells. Most of the rest of the genome is transcribed into noncoding RNA whose function is largely unknown but which is suspected of playing a variety of roles in gene regulation and evolution.
Many of these noncoding RNAs are copied from repeated DNA sequences called short interspersed nuclear elements (SINEs). Once transcribed into RNA, they can be copied back into DNA and “pasted” into the genome at new locations in a process called retrotransposition. Sometimes these new copies land in or near genes and can damage them. Other times they can add new properties to the gene.
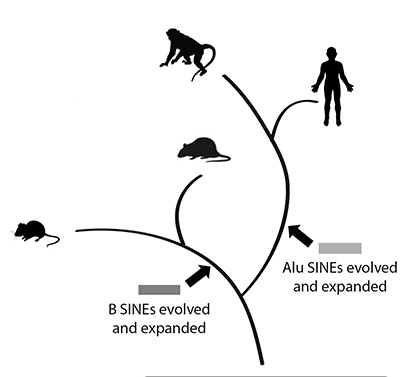
In the human genome, the major SINE family is made of so-called “Alu elements.” There are more than 1 million copies of Alu, comprising more than 10 percent of human DNA, scattered throughout the genome, and some of them are likely still able to jump to new locations.
The mouse genome, however, doesn’t have Alus; instead it has a distinct set of SINEs called B/ID elements. Different mammalian genomes have different SINEs because periodic bursts of retrotransposition by different SINEs continued to occur after the separation of different species from their last common ancestors. In the case of human and mouse, their lineages diverged about 90 million years ago.
“Surprisingly, when the mouse and human genomes were compared, the locations of SINEs were very similar, even though the SINEs themselves and the events that placed them at those locations were very different,” Ares said. “We wondered what could explain this apparent convergence of SINE insertion in two independently evolving genomes.”
Gene expression
Lynne Maquat’s lab at the University of Rochester found that SINEs that land in the part of the gene encoding the tails of the messenger RNA (called the 3-prime untranslated region or 3′-UTR) bring the mRNA under the control of a protein called Staufen, which down-regulates expression of the gene by a process called “Staufen-mediated decay” or SMD. Individual examples of SINE-mediated SMD were previously documented in both human and mouse cells, in each case with different SINEs.
Since the modern SINEs in the human and mouse genomes were not in the common ancestor, all of the changes in gene expression that depend on SINEs must have occurred after humans and mice separated, and not in their common ancestor.
“Normally we think of important gene expression control systems as having evolved long ago, but for regulation by SINE-mediated SMD, this cannot be the case,” Ares explained. “The question we asked was: Are there any cases where the same gene in mouse and human have been brought under SINE-mediated SMD, albeit using different SINEs at some point during the separate evolutionary histories of mice and men? If so, how many? And how common is convergent evolution of gene regulatory networks? How often do SINEs play a role in critically altering gene expression control?”
These questions were explored in a collaboration between the Rochester and UCSC labs, in which muscle precursor cells (myoblasts) from both organisms were analyzed to identify gene pairs (the mouse and human gene coding for the same protein) that had a SINE inserted in the mRNA tail and were under control of Staufen. Examples of such gene pairs in numbers greater than expected by chance would signal the possibility of convergent evolution of the Staufen regulatory network in myoblasts, where SMD is known to be important for gene control.
In myoblasts, the researchers were able to detect 24 genes that are both regulated by Staufen and have species-specific SINEs. This is a minimum number of potentially convergent pairs because not all genes are expressed in myoblasts, Ares noted. Additional experiments in the paper confirm the role of the SINE (by removing it and showing the mRNA becomes stable and insensitive to the presence of Staufen) for two gene pairs.
In addition to Ares and Maquat, the authors of the paper include first author Bronwyn Lucas, Hana Cho, and Keita Miyoshi at the University of Rochester; Eitan Lavi and Liran Carmel at the Hebrew University of Jerusalem; Lily Shiue and Sol Katzman at UC Santa Cruz; and Mikiko Siomi at the University of Tokyo.