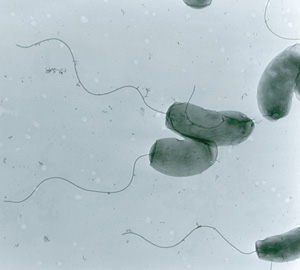
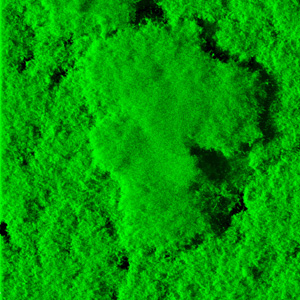
The deadly bacterium behind cholera epidemics spends only a fraction of its life infecting humans. Most of the time, Vibrio cholerae lurks in estuaries and other semisalty aquatic habitats.
Understanding the pathogen's behavior between epidemics and its transitions between different environments could help public health officials prevent outbreaks, according to microbiologist Fitnat Yildiz.
"If you know the mechanisms by which an organism can survive in the environment, you can better devise systems to prevent its transmission," said Yildiz, an associate professor of microbiology and environmental toxicology at the University of California, Santa Cruz.
The optimal salt concentration for growth and survival of V. cholerae is similar to that found in brackish water and estuaries. But the bacteria experience fluctuations in salinity both in their aquatic habitat and during human infections.
A recent analysis by Yildiz and graduate student Nick Shikuma revealed that changes in salinity affect the activity of a wide range of the pathogen's genes. Among these is a gene the researchers called OscR, which appears to play an important role in managing the pathogen's response to fluctuating salt levels in the environment. (More precisely, it responds to changes in osmolarity, a measure of the concentration of dissolved substances.) Yildiz and Shikuma described their findings in a recent paper in the Journal of Bacteriology.
Cholera outbreaks occur when environmental conditions allow the bacteria to contaminate sources of drinking water. "Cholera is a disease of disaster," Yildiz said. "Whenever there's a breakdown in public health, like during the Iraq War or in refugee camps, when people don't have access to clean drinking water, cases of cholera increase."
In areas of Africa and South Asia where cholera is endemic, outbreaks occur seasonally. Two peaks coincide with the dry season and the monsoon rains, according to Paul Dunlap of the University of Michigan, who wrote a commentary accompanying the paper in the Journal of Bacteriology. During the dry season, freshwater sources become more salty, opening them to colonization by V. cholerae. Flood conditions during monsoons can also lead to contamination of drinking water sources.
Yildiz and Shikuma found more than 300 cholera genes that respond to changes in salinity, many of which Yildiz and others had previously linked to the pathogen's deadliness. In medium salt environments similar to the human body, for instance, genes controlling virulence rev up.
The researchers found that OscR is a regulatory gene that switches on under low salt conditions and controls the activity of other genes. OscR increases bacterial motility and decreases the formation of sticky cell groupings called biofilm, Yildiz said.
Biofilm colonies form when individual cells secrete matrices of sugar chains and proteins that help them glom on to neighbors and to surfaces. Many kinds of bacteria form biofilms, which act as bacterial shields, making cells harder to kill and more tolerant of environmental fluctuations. Within biofilms, V. cholerae is more resistant to antimicrobial and chlorine-based disinfectant treatments, Yildiz said.
"What we learn with respect to the mechanisms of biofilm formation in V. cholerae could be applicable to other organisms because biofilm formation is so prevalent and it is so important for clinical, environmental, and industrial processes," she said.
V. cholerae forms biofilms both in infected hosts and in the environment. During cholera infection, biofilms cling to intestinal walls, releasing a steady stream of debilitating toxins. Scientists have even found biofilms riding through the water on tiny crustaceans called copepods, as well as in stool samples from cholera patients.
Yildiz has been investigating the network of genetic interactions that regulates biofilm formation. This network, in which environmental cues regulate genes that control other genes, works like a complex domino chain in which one piece knocks down several others.
OscR blocks the toppling dominos that lead to biofilm formation. Why the pathogen prefers being a mobile single cell in freshwater remains a mystery but is the topic of future study, Yildiz said. Knowing how the pathogen passes through freshwater versus salty water may help to expose the dynamics of cholera infections, she said.
But shifts in environmental dynamics rarely occur in isolation. Changes in salinity go hand-in-hand with changing temperatures and nutrient levels. A holistic model of V. cholerae behavior, Yildiz said, will include how these factors work in tandem.
"We are analyzing environmental fluctuations singly, but the goal is to eventually combine them," she said. "When you combine multiple stresses, you get a different response."
Yildiz collaborates with scientists in Bangladesh who help her re-create South Asian estuary or freshwater conditions in the lab. Eventually, however, she wants to study V. cholerae in the field.
Cholera is readily treated, but without treatment it causes rapid and often fatal dehydration from severe diarrhea. The World Health Organization estimates that 120,000 people die from cholera every year. Yildiz said that her work is just one step in a wide effort to predict and curb cholera epidemics.
Yildiz and Shikuma's research was funded in part by a grant from the National Institutes of Health, the ARCS Foundation, the Coastal Environmental Quality Initiative, the STEPS Institute at UCSC, and the Friends of the Long Marine Lab.