A Cancer Genomics Browser developed by researchers at the University of California, Santa Cruz, provides a new way to visualize and analyze data from studies aimed at improving cancer treatment by unraveling the complex genetic roots of the disease.
The browser consists of a suite of web-based tools designed to help researchers find patterns in the huge amounts of clinical and genomic data being gathered in large-scale cancer studies. Medical researchers hope to identify genetic signatures and other "biomarkers" in cancer cells that can be used to predict how individual patients will respond to different therapies throughout the course of their treatment.
A paper describing the Cancer Genomics Browser has been published in the April issue of Nature Methods by a team based at the Jack Baskin School of Engineering at UCSC. Coauthor David Haussler, professor of biomolecular engineering, said development of the browser was driven by the needs of cancer researchers, who are now using powerful technologies for genome analysis and DNA sequencing in their efforts to understand cancer at the molecular level.
"Each of these tests gives millions of measurements, and the result is a bad case of data overload," Haussler said. "We've built the cancer browser so that researchers can upload their data and use a variety of software tools to visualize and interpret their results."
To get a user's perspective on the browser as it took shape, Haussler's team worked closely with Dr. Laura Esserman, professor of surgery and radiology at UC San Francisco, and Marc Lenburg, associate professor of pathology and laboratory medicine at Boston University School of Medicine. Esserman and Lenburg, both coauthors of the paper, are involved in the I-SPY Trial, a multi-institutional collaboration aimed at identifying biomarkers to predict the most effective therapies for patients with advanced breast cancer.
"What is amazing about the browser is that it allows us to combine complex molecular data and clinical observations, and provides insights into how we can truly improve treatment and outcomes," said Esserman, director of the Carol Franc Buck Breast Care Center and associate director of the Breast Oncology Program at the Helen Diller Family Comprehensive Cancer Center at UCSF.
Cancer genomics involves searching for all of the genes and mutations that contribute to the development of a cancer cell and its progression from a localized cancer to metastatic disease that spreads throughout the body. A genome is an organism's complete set of DNA, and researchers are now able to analyze the alterations that occur throughout the genome of a patient's cancer cells. Recent advances, such as microarray technology and high-throughput DNA sequencing, have made it possible to characterize tumor samples in exquisite detail.
"You can run a microarray chip that analyzes a million points in the genome and can tell you about changes in the DNA, as well as inherited variations that make a person more or less susceptible to cancer," Haussler said.
Many different types of genomic changes can have clinical significance, including insertions, deletions, and other changes in the DNA sequence, such as changes in the number of copies of a gene. Moreover, microarrays and high-throughput methods for measuring proteins make it possible to see how these genomic alterations interfere with the cell's normal workings.
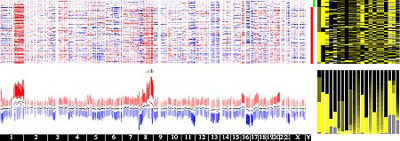
"The Cancer Genomics Browser is fantastic in that it helps users display many different dimensions of clinical and molecular data simultaneously," Lenburg said. "For example, for a given set of tumor biopsies, it is possible to see which regions of the genome are abnormal, how much of every gene is being expressed, how active various signaling pathways are--all organized by, say, how well each patient responded to a particular drug. As a result, the process of identifying possible connections is really easy."
The browser was developed by a team of scientists at UCSC's Center for Biomolecular Science and Engineering (CBSE), an interdisciplinary center housed in the Baskin School of Engineering and directed by Haussler. Ting Wang, a Helen Hay Whitney postdoctoral fellow, came up with the initial design of the browser and coordinated the team's efforts. The first three authors of the paper--postdoctoral researcher Jingchun Zhu and graduate students Zachary Sanborn and Stephen Benz--did much of the work involved in building the browser, with help from CBSE research scientist James Kent and others.
The public browser site hosts a growing body of publicly available cancer genomic data, and the browser is also being used on confidential, prepublication data by several groups involved in clinical trials and cancer genomics research, Wang said.
The Cancer Genomics Browser is a natural extension of the UCSC Genome Browser, a widely used platform for accessing and visualizing genomic data. Created by Kent as a tool for exploring the human genome, the UCSC Genome Browser now averages one million page requests every week. It displays data and annotations in linear tracks that parallel the DNA sequences of the dozens of genomes in the browser.
But this type of display doesn't work well with clinical data from large numbers of patients. And clinical databases don't handle genomic data very well. The Cancer Genomics Browser is able to integrate these different types of data into a single interactive display.
"Large clinical trials that include detailed molecular profiling of patient samples generate a really big mountain of data. Actually, it is more like several big mountains of data," Lenburg said. "The browser creates a way of organizing all this data, and all these different types of data, into a single unified picture."
The Cancer Genomics Browser represents data as "heatmaps," in which colors represent the values of key variables. Genomic and clinical data are displayed side by side, and researchers can group and sort the data on the basis of any feature of interest, such as age, gender, response to therapy, estrogen-receptor status of breast cancers, and so on. Because humans excel at visual pattern recognition, correlations in the data tend to jump out as the user manipulates the browser display.
"The ideas behind it are simple, but the result is a pretty powerful tool. It makes it a lot easier to see patterns in the data," Wang said.
Standard statistical tools are integrated into the browser so that users can perform quantitative analyses. The browser's developers hope to improve these capabilities in the future. "Now that we have the platform, we want to incorporate state-of-the-art algorithms to get the most out of the data," Wang said.
In developing the browser, the researchers used prepublication datasets from the I-SPY Trial (Investigation of Serial Studies to Predict Your Therapeutic Response with Imaging and Molecular Analysis) and The Cancer Genome Atlas (TCGA). The I-SPY study is funded by the National Cancer Institute (NCI) and includes nine cancer centers nationwide. TCGA is a large-scale collaborative effort by NCI and the National Human Genome Research Institute to systematically characterize the genomic changes that occur in cancer. The UCSC team is also working with a related worldwide effort, the International Cancer Genome Consortium.
The coauthors of the Nature Methods paper include UCSC researchers Christopher Szeto, Fan Hsu, Robert Kuhn, Donna Karolchik, and John Archie, in addition to Zhu, Sanborn, Benz, Lenburg, Esserman, Kent, Haussler, and Wang. Funding for this project was provided by the I-SPY consortium, the TCGA consortium, the California Institute for Quantitative Biosciences (QB3), and the National Institutes of Health. Haussler is a Howard Hughes Medical Institute investigator.