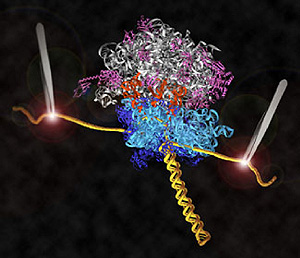
Research by UC Santa Cruz molecular biologist Harry Noller and his collaborators has led to the first direct observations of the mechanism for protein synthesis in living cells. Their new findings on ribosomes, the protein-making molecular machines in all cells, are featured on the cover of the April 3, 2008, issue of the international science journal Nature.
The researchers used a laser apparatus called "optical tweezers" to probe the physical steps of the ribosome machine as it translates genetic code into a protein molecule.
Noller, the Sinsheimer Professor of Molecular Biology at UCSC, has been studying the ribosome for more than 30 years. His latest findings are the result of a six-year collaboration among three groups within the California Institute for Quantitative Biosciences (QB3). The joint effort involved Noller and postdoctoral researcher Laura Lancaster at UCSC and the UC Berkeley labs of biophysicist Carlos Bustamante and biophysical chemist Ignacio Tinoco, Jr.
"This is a collaboration between the tweezers guys at Berkeley and the ribosome guy at UCSC," Noller said. "We got together to figure out if we could use this approach to measure forces exerted by ribosomes during protein synthesis."
To make a new protein, the genetic instructions are first copied from the DNA sequence of a gene into a messenger RNA molecule. The ribosome then reads the genetic code from the messenger RNA and translates the code into the structure of a protein.
The Noller lab engineered ribosomes containing messenger RNA molecules with strands of DNA attached to both ends to serve as "handles." The DNA strands, in turn, are attached to tiny beads. Both beads are fixed by lasers in the optical-tweezers apparatus, and the lasers at each end exert opposing forces on the translation system.
Using this system, the researchers were able to follow the translation process in individual ribosomes. They found that protein synthesis has a beat: bop, bop, bop, pause; bop, bop, bop, pause; and so on. The three "bops" correspond to the ribosome reading one "codon"--a sequence of three RNA subunits that tell the ribosome to add a specific amino acid to the protein chain.
"The ribosome moves along the message in a series of stop-and-go events--stalling and translocation. Pause, translocate, pause," Noller said.
Noller's research group was the first to solve the complete structure of a ribosome using x-ray crystallography. That was the first step in answering a bigger question: How does the ribosome operate?
"Until now, we were limited to watching a few trillion ribosomes, and they were not synchronized--the details were blurred. Now we can watch one ribosome at a time," he said.
Noller, who directs the Center for Molecular Biology of RNA at UCSC, said the next phase in this project involves refining the analysis. "We have not yet directly measured the forces exerted by the ribosome," he said. "Now we have a more sensitive tweezers setup at UC Berkeley, and we have devised a way of grabbing a ribosome with one tweezers and the message with the other. We can feel the ribosome pulling on the message."
The first author of the Nature paper is Jin-Der Wen of UC Berkeley. In addition to Noller, Lancaster, Bustamante, and Tinoco, the coauthors include Courtney Hodges of UC Berkeley; Ana-Carolina Zeri of the Brazilian Synchrotron Light Laboratory; and Shige Yoshimura of Kyoto University.
The California Institute for Quantitative Biosciences (QB3) is a cooperative effort among three UC campuses--Santa Cruz, Berkeley, and San Francisco--and private industry.