Earth & Space
Scientists find unexpected proteins in bacteria motors
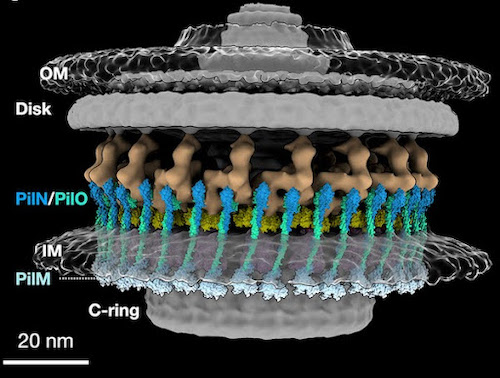
A team of scientists, co-led by Karen Ottemann, a professor of Microbiology and Environmental Toxicology, recently found three unexpected proteins while studying the motors that power the flagella of a species called Helicobacter pylori. The proteins, which are normally found in another type of appendage on a separate group of bacteria, seem to exert control over the motion of the flagella. These proteins, known as PilN, PilO, and PilM, had never been found associated with a flagella before.
How well bacteria move and sense their environment directly affects their success in surviving and spreading. About half of known bacteria species use a flagella to move — a rotating appendage that functions like a propellor. The flagella have motors behind them with tiny cylinders that look almost mechanical.
A team of scientists recently found three unexpected proteins while studying the motors that power the flagella of a species called Helicobacter pylori. The proteins, which are normally found in another type of appendage on a separate group of bacteria, seem to exert control over the motion of the flagella. These proteins, known as PilN, PilO, and PilM, had never been found associated with a flagella before.
“They’d always been found in a totally different type of motor organelle called pili that don’t rotate. Pili extend and retract more like Spiderman grappling hooks. So nobody thought that flagella and pili would have parts in common because they’re so different,” said Karen Ottemann, a professor in the Department of Microbiology and Environmental Toxicology at the University of California, Santa Cruz, and one of the lead authors on the Proceedings of the National Academy of Sciences paper that describes the results.
While talking with another professor in the field, Ottemann realized both labs had made the same discovery using different methods, “just by chance. And because we’re friendly and a collaborative field, we actually talk to each other,” she said.
After that conversation, Ottemann helped form a collaboration between labs at three different universities. The international team soon began teasing apart how PilM, PilN and PilO affect the behavior of the bacteria.
Ottemann’s lab focused on the biology and molecular genetics of H Pylori. The Liu lab at Yale University used cryo-electron tomography and microscopy techniques to provide detailed images of the motors inside the bacteria. The Roujeinikova lab at Monash University in Australia focused on biochemistry and studied how PilO, PilN and Pill interact with each other and the flagellar motor.
The scientists hypothesized that H Pylori bacteria with larger, more complex flagella that contained the proteins would outperform bacteria with flagella that lacked them. They removed the genes individually and in combination, and to their surprise, the proteins did not increase the speed or torque of the flagella.
“We were really puzzled by that at first because when we got rid of the proteins the bacteria actually migrated better,” said Ottemann. This stumped the lab until UC Santa Cruz postdoc Xiaolin Lium made careful observations of the bacteria while they were migrating. The scientists realized that H pylori with the proteins stop and wait before moving when they encounter a new, thicker medium. The bacteria without the proteins forge ahead without pause.
“Instead of making the flagella rotate more strongly, those parts seem to instead confer some kind of control on it,” said Ottemann. “The bacteria want to stop for some reason still unknown to us.”
Bacteria don’t have genes they don’t need, Ottemann explains. “So they clearly need this ability to stop.” One of the group’s next challenges will be to figure out why and how the proteins stop the flagella motors.
“We don’t actually know whether the flagella are fully stopped, or if they’re rotating but kind of disconnected from the motor — like a brake versus a clutch,” Ottemann said.
The three labs will continue their collaboration for the next steps.
“You need diverse viewpoints to tackle a complex new problem like this,” she said. “I like working with this team, and there are still some good questions to ask.”