Campus News
New high-throughput approach yields libraries of probes for immunological assays
Technical innovation enables rapid assessment of T cell repertoires for research and diagnostics, with potential applications in studying COVID-19 immune responses.
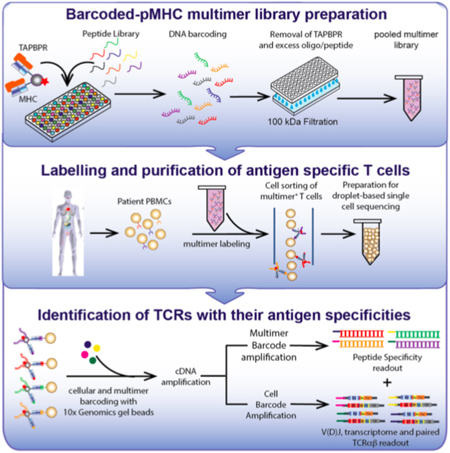

An immunological test known as a “tetramer assay” can detect and quantify the T cells in a blood sample that are able to recognize a specific antigen, such as a viral protein. Making the molecular probes needed for this type of assay, however, has always been a difficult and time-consuming process.
Now, a team led by researchers at UC Santa Cruz has developed a method to create libraries of probes for high-throughput, large-scale assessments of T cell repertoires in blood samples. The new approach, described in a paper published April 20 in Nature Communications, opens up new opportunities for immunological research, development of cancer immunotherapies, and assessing the immune responses of patients with viral infections, including COVID-19.
“Every person in the field knows how cumbersome it is to make the probes for these assays,” said corresponding author Nikolaos Sgourakis, assistant professor of chemistry and biochemistry at UC Santa Cruz. “With conventional methods, you would need about a week to make a single peptide complex, but now we can make a plate of 100 in a day.”
T cells play a central role in the immune system, complementary to antibodies (produced by B cells). Antibodies recognize and bind to antigens (foreign proteins) in the blood and other body fluids, whereas T cells only bind to antigens displayed on the surfaces of cells in the body, enabling the immune system to detect infected or cancerous cells.
This difference makes antibody tests relatively straightforward and T cell assays much more challenging. To construct a probe for detecting a specific T cell receptor, the corresponding antigen must be incorporated into a molecular complex that mimics the way antigens are displayed on cell surfaces, bound to major histocompatibility complex (MHC) proteins. Sgourakis has been studying how protein fragments are selected and bound to MHC proteins in cells, and the new method builds on his lab’s discoveries about the role of “molecular chaperones” in this process.
“Molecular chaperones are designed by nature to load MHC proteins with peptides in the cell, so we took our favorite chaperone and repurposed it,” Sgourakis said.
His previous research had shown that the chaperone can eject antigens that have low affinity for the MHC protein, ensuring that it binds only high-affinity antigens that can be displayed at the cell surface in the proper conformation to activate a T cell response. So Sgourakis designed a “placeholder” peptide for use in preparing large quantities of pre-loaded MHC complexes. When incubated with a high-affinity antigen, the placeholder is displaced, and this reaction can be performed in parallel with large numbers of antigens in a high-throughput system.
“It’s a force multiplier, enabling us to perform these reactions at high throughput,” Sgourakis said. “A lot of groups are working on similar methodologies, all of which have their pros and cons. This technology has the advantage of using the same system that cells use naturally, and we can combine it very elegantly with existing single-cell analytical tools.”
This work was done in close collaboration with researchers at the New York Genome Center and the University of Pennsylvania’s Children’s Hospital and Perelman School of Medicine. The researchers started to use the new method to develop libraries of probes for assessing T cell responses to neuroblastoma and designing cancer immunotherapies.
Then came COVID-19, and the team began exploring ways to apply the new technology to address the challenges of the novel coronavirus. With a viral infection, there are many different fragments of the viral proteins that an infected cell can display, and it is important to determine which of these peptides elicit a strong immune response.
“Based on the coronavirus genome, we can predict all the possible peptides, synthesize them, load them onto MHC tetramers, and do a fishing expedition to find which ones are recognized by the T cells in blood samples from patients,” Sgourakis explained. “Certain peptides are immunodominant—they steer the immune response—and those are the ones we want to discover so we can potentially use them in a vaccine.”
This approach can also be used to compare the T cell receptor repertoires in different cohorts of patients. As people age, their T cell repertoire declines, resulting in a diminished ability to mount an immune response to a novel threat. This may be why older people are more vulnerable to COVID-19.
“One of the big questions is why there is so much variability in the severity of this disease,” Sgourakis said. “We can use this technology to screen patients and see what the gaps are in their T cell repertoires, and maybe use this as a diagnostic for which patients will need more intensive treatment.”
The coauthors of the paper include co-first authors Sarah Overall and Jugmohit Toor at UC Santa Cruz; Stephanie Hao and Peter Smibert at the New York Genome Center; Mark Yarmarkovich, Son Nguyen, Alberto Japp, Michael Betts, and John Maris at the University of Pennsylvania; and Sara O’Rourke, Giora Morozov, Nicolas Gonzalez, and Danai Moschidi at UC Santa Cruz. This work was funded by the National Institutes of Health.