Campus News
Genes found only in humans influence brain size
A set of three nearly identical genes found only in humans appear to play a critical role in the development of our large brains, according to a study led by UCSC researchers.


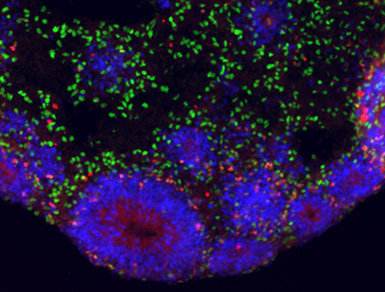
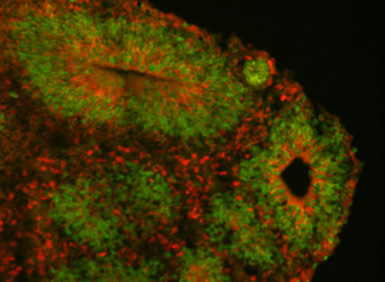
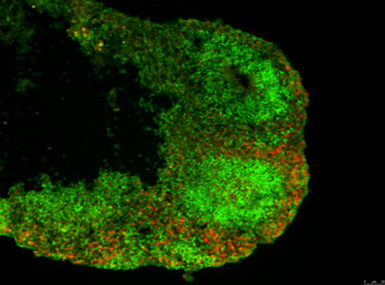
A set of three nearly identical genes found only in humans appear to play a critical role in the development of our large brains, according to a study led by researchers at UC Santa Cruz.
The genes appeared between 3 and 4 million years ago, just before the period when fossils show a dramatic increase in the brain sizes of human ancestors. In modern humans, the genes are involved in genetic defects associated with neurological disorders.
Published May 31 in Cell, the study represents more than five years of work to characterize the genes, their role in neurological development, and their evolutionary origins. They belong to an ancient family of genes known as Notch genes, first discovered in fruit flies and named for a genetic defect causing notched wings.
“This is a family of genes that goes back hundreds of millions of years in evolutionary history and is known to play important roles in embryonic development. To find that humans have a new member of this family that is involved in brain development is extremely exciting,” said senior author David Haussler, professor of biomolecular engineering and scientific director of the UC Santa Cruz Genomics Institute.
The site of the genes on the long arm of chromosome 1 is involved in genetic defects in which large segments of DNA are either duplicated or deleted, leading to neurological disorders known collectively as 1q21.1 deletion/duplication syndrome. Deletions are often associated with microcephaly (abnormally small head size) and autism, while duplications are often associated with macrocephaly (abnormally large head size) and schizophrenia.
Common ancestor
The new human-specific Notch genes were derived from NOTCH2, one of four previously known mammalian Notch genes, through a duplication event that inserted an extra partial copy of NOTCH2 into the genome. This happened in an ancient ape species that was a common ancestor of humans, chimpanzees, and gorillas. The partial duplicate was a nonfunctional “pseudogene,” versions of which are still found in chimp and gorilla genomes. In the human lineage, however, this pseudogene was “revived” when additional NOTCH2 DNA was copied into its place, creating a functional gene. This new gene was then duplicated several more times, resulting in four related genes, called NOTCH2NL genes, found only in humans.
One of the four NOTCH2NL genes appears to be a nonfunctional pseudogene, but the other three (NOTCH2NLA, NOTCH2NLB, and NOTCH2NLC) are active genes that direct the production of truncated versions of the original NOTCH2 protein. Notch proteins are involved in signaling between and within cells. In many cases, the Notch signaling pathway regulates the differentiation of stem cells in developing organs throughout the body, telling stem cells when to become, for example, mature heart cells or neurons.
“Notch signaling was already known to be important in the developing nervous system,” said senior author Sofie Salama, a research scientist at UC Santa Cruz. “NOTCH2NL seems to amplify Notch signaling, which leads to increased proliferation of neural stem cells and delayed neural maturation.”
Radial glia
The NOTCH2NL genes are especially active in the pool of neural stem cells thought to generate most of the cortical neurons. By delaying their maturation, the genes allow a larger pool of these stem cells (called “radial glia”) to build up in the developing brain, ultimately leading to a larger number of mature neurons in the neocortex (the outer layer of the brain in mammals; in humans, it hosts higher cognitive functions such as language and reasoning).
This delayed development of cortical neurons fits a pattern of delayed maturation characteristic of human development, Haussler said. “One of our most distinguishing features is larger brains and delayed brain development, and now we’re seeing molecular mechanisms supporting this evolutionary trend even at a very early stage of brain development,” he said.
Salama noted that the new genes are just one of many factors that contribute to cortical development in humans. “NOTCH2NL doesn’t act in a vacuum, but it arose at a provocative time in human evolution, and it is associated with neural developmental disorders. That combination makes it especially interesting,” she said.
The DNA copying errors that created the NOTCH2NL genes in the first place are the same type of errors that cause the 1q21.1 deletion/duplication syndrome. These errors tend to occur in places on the chromosomes where there are long stretches of nearly identical DNA sequences.
“These long segments of DNA that are almost identical can confuse the replication machinery and cause instability in the genome,” Haussler explained. “We may have gained our larger brains in part through the duplication of these genes, but at the expense of greater instability in that region of chromosome 1, which makes us susceptible to the deletion/duplication syndrome.”
Reference genome
Long stretches of repetitive DNA also present challenges for DNA sequencing technologies. In fact, the location of NOTCH2NL in the human reference genome was not accurate when Haussler’s team first started investigating it.
“When we looked at the reference genome to see where NOTCH2NL was, we found that it was near the area involved in the 1q21.1 syndrome, but not part of the region that was deleted or duplicated,” Haussler said. “This explains why the gene was not looked at before by geneticists studying the syndrome.”
After checking other genome data and contacting the team working on the next iteration of the reference genome, however, Haussler found that NOTCH2NL is in fact located in the interval where the defects occur. The new reference genome (the 38th version, released later in 2013) also shows the additional copies of the gene. Haussler’s team subsequently showed that the duplications or deletions in the syndrome result in an increase or decrease (respectively) in the number of copies of NOTCH2NL genes in the affected person’s genome. Other genes are also duplicated or deleted and may also be involved in the syndrome.
Interestingly, these genetic changes do not always result in neurological disorders. In about 20 to 50 percent of affected children, the syndrome is the result of a new genetic mistake, but in many cases one of the parents is found to also carry the genetic defect, without showing any apparent symptoms. According to Haussler, this is not uncommon in genetic diseases and underscores the importance of multiple factors in the development of disease.
“It’s amazing how often we find people with what seem to be serious genetic conditions, yet something else compensates for it,” he said.
Cortical organoids
The investigation of these genes began in 2012 when Frank Jacobs, now at the University of Amsterdam and the third senior author of the paper, was working with Haussler and Salama at UC Santa Cruz as a postdoctoral researcher. His project involved coaxing human embryonic stem cells to differentiate into neurons and studying the genes that are expressed during this process. As the cells develop into cortical neurons in the petri dish, they self-organize into a layered structure like a miniature version of the brain’s cortex, which researchers call a “cortical organoid.”
Jacobs was comparing gene expression patterns in cortical organoids grown from embryonic stem cells of humans and rhesus monkeys. Many genes showed differences in the timing and amount of expression, but NOTCH2NL was exceptional. “It was screaming hot in human cells and zero in rhesus. Rhesus cells just don’t have this gene,” Salama said. “Finding a new Notch gene in humans set us off on a long journey.”
Haussler, a Howard Hughes Medical Institute investigator, said he remembers presenting their initial findings in 2013 to scientists at HHMI. “Their general reaction was, ‘Well, it’s amazing if it’s true, but we’re not convinced yet.’ So we spent the next five years working to convince everybody.”
The development of the CRISPR/Cas9 system for making genetic modifications provided a crucial tool for their work. Salama’s team used it to delete the NOTCH2NL genes from human embryonic stem cells. Cortical organoids grown from these cells showed an acceleration of neural maturation and were smaller in size than organoids from normal cells. The researchers also inserted NOTCH2NL genes into mouse embryonic stem cells and showed that the genes promote Notch signaling and delay neural maturation in mouse cortical organoids.
“The fact that we can genetically manipulate stem cells with CRISPR and then grow them into cortical organoids in the lab is extremely powerful,” Haussler said. “My dream for decades has been to peer into human evolution at the level of individual genes and gene functions. It’s incredibly exciting that we’re able to do that now.”
Modern humans
A major part of the research involved careful and precise sequencing of the region of chromosome 1 where the NOTCH2NL genes are located in 8 normal individuals and 6 patients with 1q21.1 deletion/duplication syndrome. (The researchers also analyzed the genomes of three archaic humans, two Neanderthals, and one Denisovan, finding in all of them the same three active NOTCH2NL genes that are present in modern humans.)
The sequencing results showed that the NOTCH2NL genes are variable in modern humans. The researchers identified eight different versions of NOTCH2NL, and Haussler said there are probably more. Each version has a slightly different DNA sequence, but it remains unclear what effects these differences may have.
“We’ve found that all of them can promote Notch signaling. They behaved in subtly different ways when we tested them in cell cultures, but we have a lot more work to do before we can start to get a handle on what this means,” Salama said.
Other genes involved in human brain development seem to have arisen through a duplication process similar to the creation of NOTCH2NL. A notable example is the gene SRGAP2C, which is thought to increase the number of connections between neurons. Locations in the genome where such duplications and rearrangements occur repeatedly, known as “duplication hubs,” make up about 5 percent of the human genome and seem to have been important in human evolution, Haussler said.
The first authors of the paper are Ian Fiddes, a graduate student working with Haussler at UC Santa Cruz, and Gerrald Lodewijk, a graduate student working with Jacobs at the University of Amsterdam. Other coauthors include researchers at Stanford University, UC San Francisco, University of Washington, Broad Institute of MIT and Harvard, Medical Genetics Service in Lausanne, Switzerland, and Institute of Genetic Medicine in Newcastle upon Tyne, U.K. This work was supported by the Howard Hughes Medical Institute, U.S. National Institutes of Health, European Research Council, California Institute for Regenerative Medicine, Netherlands Organization for Scientific Research (NWO), and European Molecular Biology Organization.